Contribute
| Technology - High-Throughput Genome Sequencing |
Shailendra Yadav
03/14/2005
A genome of an organism is the collection of all the genes in the
organism’s complete set of chromosomes. Genes, which are units of
heredity, encode all the proteins responsible for controlling
everything from organism’s shape and size down to the function of every
organ, tissue and cell. On a molecular level, genes are ordered
sequence of four letters: A, C, G and T, which represent four
nitrogenous-rich chemicals, also called bases, held together by weak
bonds such that A pairs only with T and G pairs only with C. The
long chain consisting of two strands that result from these paired
bases form a double-helix structure, called deoxyribonucleic acid (DNA)
(Fig: 1). A human genome consists of approximately 3 billion base
pairs, which make up about 30,000 genes. Genome sequence, hence, is
essentially a read-out of the entire sequence of the base pairs.
So how do you obtain the sequence of a genome? Genome sequencing
technology has evolved considerably from the days of extremely
low-throughput, high-cost and labor-intensive slab gel based sequencing
to the current day state-of-the-art high-throughput, lower-cost and
automated capillary-electrophoresis based sequencing done at Broad
Institute, Cambridge, MA (www.broad.mit.edu). Our sequencing platform
can currently sequence a human genome within 3 months at a cost of
under 10 million dollars. The Human Genome Project, the first such
effort to sequence a complete human genome completed in 2002 and took
11 years and 1 billion dollars.
The genome shotgun sequencing
is based on the Sanger sequencing method (named after British scientist
Fred Sanger, who invented the method in 1977). Due to the limitation of
current technology, you can’t just put a whole genome through a
sequencer and directly read the sequence of that genome. First you take
the genome and cut it into hundreds of thousands of small unique
fragments of overlapping sequence, each about a few thousands base
pairs long. Next you need to make enough (on the order of tens of
millions) copies of each fragment, attach fluorescent label through
enzymatic reaction and run them through capillaries, where the labels
from all the fragments collectively fluoresce well enough to be
detected by a laser.
Before getting copied, DNA fragments
are first attached to cloning vectors, which are circular pieces of DNA
into which the fragment to be copied is introduced. This step is called
template preparation. Next, these vectors with attached fragments are
introduced into bacteria called E. Coli., which under the right
conditions of temperature, time and nutrition multiply exponentially
together with the vectors inside it. Thus, you end up with millions of
copies of each fragment. In a high-throughput laboratory environment,
each of the colonies of these fragments, also called clones, is picked
into different wells of 384-well plate pre-filled with growth media,
using a 3-axis robot ( Fig: 2) with 384 pins that are decontaminated
between plates. For a single mammalian genome, there are about tens of
thousands of these plates that need to be processed over few months.
The plates are fed through automated stackers on the robot that
processes hundreds of plates per day. After the bacterial growth step,
the plates are through another robotic platform, where all the clones
are transferred into another special 384-well plate capable of
withstanding higher temperature for the enzymatic reaction to occur.
Together with clones, an enzyme called phi29 is added into the plates.
All the plates have unique barcodes which are tracked in real-time
through a Laboratory Information Management System (LIMS). These robots
are capable of processing up to 1200 plates/day (Fig: 3). The automated
liquid dispenser unit with 384 disposable pipette tips mounted on this
robot has the capacity to deliver sample volume as low as a few hundred
nanoliters into each well. Low volume range is a critical requirement
for the dispenser to accomplish lowest possible usage of the highly
expensive enzymes and reagents used for the reactions in this and the
sequencing step. The plates are then incubated at room temperature for
about 16 hours during the amplification step where bacterial cells are
lysed to expose the inserted genomic DNA, which is then exponentially
amplified based on rolling circle amplification. This step results in
enough DNA for the sequencing reaction to occur in the next step.
In the sequencing step, DNA is transferred into another plate with the
addition of another enzyme called DNA polymerase together with four
different colored fluorescent labels representing the four different
bases. This most expensive reagent is added using a special non-contact
dispensing robot that dispenses on-the-fly. This dispenser is capable
of dispensing on the order of tens of nanoliters. The plates are then
heat cycled resulting in sequencing products with fluorescent labels
attached to them. Finally, these plates are put on the detectors (Fig:
4), where the products are electrokinetically injected into
capillaries, which have been filled with the separation matrix. DNA,
being negatively charged, moves toward the positive end of the
capillaries upon the application of electrical voltage. The speed of
the migration is a function of DNA size. So by taking into account the
speed and the dye color (unique for each base), detected through the
laser, the software identifies the sequence of bases on a given
fragment. Repeating this process for all the fragments, you finally
have the sequence (Fig: 5) of all the small fragments of DNA. Since all
of these fragments have overlapping sequences, the next step is to
assemble them into whole genome based on the overlapping ends.
Various other technologies are being developed at Broad and elsewhere
with the goal of bringing the cost of sequencing one mammalian genome
down, first to $ 100,000 and then to $ 1000, while cutting
time down, first to weeks and then to days. The sequencing
community is actively developing other platforms (microfluidic chip
with offline detection, integrated microfluidic and online optical
detection etc) as well as pursuing other approaches (single molecule
sequencing using zero-mode waveguides, solid-state nanopores etc) in
the hope of reaping the benefits promised by these technologies. Only
time will tell us which technology is best suited to advance our
ability to sequence genome quickly, cheaply and effectively.
You may also access this article through our web-site http://www.lokvani.com/


Fig 1: DNA

Fig 2: Colony Picking Robot

Fig 3: High throughput Laboratory Automation System

Fig 4: Sequence Detectors
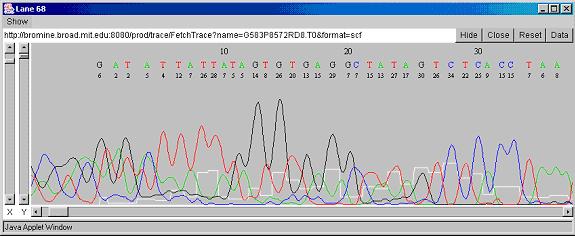
Fig 5: DNA Sequence