Contribute
| Technology - Macromolecular Crystallography: Its Role In Understanding Disease And In Drug Development |
Chaitanya Hiremath
03/12/2004
(This article is sponsored by The Boston Group)
Introduction
Proteins, often described as workhorses and nanomachines, are essential constituents of living cells. Over 30,000 different proteins are synthesized by the mammalian cell, which work in harmony to carry out a great variety of tasks, from providing the structural scaffolding for our cells, tissues, muscles, and bones, to serving as hormonal messengers and enzymes, regulating biochemical processes, and relaying genetic information, among other duties. Proteins are made up of molecular units called amino acids, whose sequence is dictated by DNA. Their specialized and powerful biochemical capabilities are solely due to their unique shapes.
A detailed understanding of the relationship between molecular structure and function is one of the fundamental aims of modern biology. Information on the structure can be obtained by physical methods such as sedimentation behavior, electron microscopy, neutron and X-ray low angle scattering, nuclear magnetic resonance (NMR) and X-ray crystallography. X-ray crystallography is arguably the most effective of these techniques at present. Certainly the other techniques complement crystallography and have a valued place in the set of tools available. In contrast to NMR, which is an indirect spectroscopic method in solution, no size limitation exists for the molecule or complex to be studied. X-ray crystallography is instrumental in studying the three-dimensional structure of macromolecules such as DNA, RNA, proteins, ribosomes and viruses. Advances in crystallography have been central to the explosive development of the area of structure-function relationships for biomolecules.
What is Crystallography?
Can we see atoms using a microscope? Using visible light, it will never be possible to see atoms under even the most powerful of microscopes. In order for an object to be seen, its size needs to be at least half the wavelength of the light being used to see it. Since visible light has a wavelength in the range of 4000-7000 Ångstrom (10-10 m) and the size of atoms is about 1 Å, it is not possible to see molecules using visible light. Are there smaller wavelengths of light that can be used to see atoms? X-rays are electromagnetic radiation of wavelength about 1 Å (10-10 m), which is comparable to the size as an atom and the distance between atoms in a molecule. The discovery of X-rays in 1895 enabled scientists to probe crystalline structure at the atomic level.
X-ray -> Crystal -> Diffraction Pattern -> Fourier Transform -> Electron Density -> 3-D Structure
The first and the most important step in X-ray crystallography is growing a pure crystal of the molecule whose structure is to be determined. In a crystal, the molecule is repeated throughout the crystal in all directions. When a finely focused beam of X-rays is passed through the crystal, X-rays are deflected only in certain definite directions producing a diffraction pattern. A diffraction pattern is a complex pattern of spots with different intensities, which can be recorded on a photographic film or an electronic detector. Encoded in the pattern is information about the positions of the atoms in the crystal. Using the mathematical Fourier transform these patterns are converted into electron density maps, displayed as contour maps resembling topographic maps in geography. The peaks in the electron density map correspond to the atomic positions in the molecule. Finally, from that map a three-dimensional model of the molecule is constructed.
Structural biology had its first triumphs in 1950s, resulting in a series of Nobel prizes. Linus Pauling’s postulation of the alpha-helix and beta-sheet as central features of protein structures was followed by the Watson-Crick proposal of the double helical nature of DNA (1953). By 1954, Dorothy Hodgkin at Oxford had solved the structure of vitamin B12. In the late 1950s, Kendrew and Perutz at Cambridge had obtained the first electron density maps of proteins myoglobin and oxyhemoglobin. Several hundred unique structures have been determined so far. Knowledge of 3-dimensional structures of various biomolecules has had a profound impact on the whole of biology.
Recent advancement
Modern X-ray crystallography is a multi-disciplinary field that provides the most powerful and accurate method for determining single-crystal structures. Structures containing 100–200 atoms now can be analyzed on the order of 1–2 days, whereas before the 1960s a 20-atom structure required 1–2 years for analysis. Technological advancements have been achieved in all aspects of crystallography. The most significant technical advancement has been the impressive increase in the quality and quantity of the samples purified by using molecular biology and biochemical techniques. Flash cooling protein crystals to cryogenic temperatures (~100 K) offers many advantages, the most prominent of which is the elimination of radiation damage. Synchrotron radiation, high-intensity and high-energy X-ray source, has revolutionised structural biology by providing state-of-the-art facilities and methods. These reliable and tunable sources, which considerably reduce the exposure time from hours to seconds, are available at several places around the world, such as Cornell High Energy Sychrotron Source (CHESS) at Cornell University and National Sychrotron Light Source at Brookhaven National Laboratory in New York, among others. Diffraction experiments are greatly aided by the use of computer-controlled diffractometers and powerful data analysis software. Inferring the atomic structure from the diffraction data has required the development of complex mathematical methods using Fourier Transforms, manipulation of 3D electronic density data, Monte-Carlo and genetic algorithms, making Crystallography a field of research hungry for computing power and new algorithms. Advances in computer technology have made it possible to tackle more complex structures such as viruses (Figure 1).
The state-of-art macromolecular crystallography has given us new eyes to look at biology. A comprehensive understanding of biochemical and cellular processes is possible from the three-dimensional atomic structures of proteins and other biological macromolecule. Drug researchers experimentally determine the structure of a target, if possible with a bound inhibitor, and use the structural information to guide the synthesis of new compounds based on size, shape and chemical and physical properties that lead to optimal binding to target. The knowledge of accurate molecular structures is a prerequisite for rational drug design and for structure based functional studies to aid the development of effective therapeutic agents and drugs.
(Chaitanya Hiremath, Ph.D, is a Structural Biologist. He was a Senior-Fellow at Harvard Medical School and has worked in the biopharmaceutical industry. He has contributed a chapter on “Structure and Assembly of Viruses” in the book “Molecular Conformation and Biological Interactions” edited by P. Balaram and Ramaseshan (Indian Academy of Sciences, 1991) and has published several papers in peer-reviewed journals. )You may also access this article through our web-site http://www.lokvani.com/

Chaitanya Hiremath
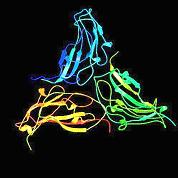
Figure (a)

An example of a virus structure. Large number of identical protein subunits (a) form a highly symmetrical protein shell (b) that encapsulates the genome.
